Prediction of secreted proteins
Overview
Teaching: 10 min
Exercises: 30 minQuestions
How can I predict proteins that are secreted?
How do I get my analysis results back to my computer?
Objectives
Generate a list of potential secreted proteins.
Transfer data out of a cloud session.
Protein secretion prediction
SignalP is a neural network–based method which can discriminate signal peptides from transmembrane regions. A signal peptide is the N-terminal part of a protein that is targeted to the secretory pathway in pro- and eukaryotes. In prokaryotes, translocation takes place across the cytoplasmic membrane (inner membrane in Gram-negative bacteria), and the process can happen during or after translation.
However the presence of a signal peptide does not necessarily mean that the protein is secreted to the extracellular environment—it only means that it enters the secretory pathway. The protein could have one or more transmembrane helices downstream of the signal peptide and therefore be retained in the membrane. In Gram-negative bacteria, the protein could be retained in the periplasm, or be inserted into the outer membrane as a β-barrel transmembrane protein. In Gram-positive bacteria, the protein could be attached to the cell wall. In general signal peptides have three regions:
- an N-terminal n-region of variable length characterized by positive charge
- a central h-region of at least 7 hydrophobic residues\
- a C-terminal c-region of typically 3-7 polar residues.
Positions –1 and –3 relative to the cleavage site are occupied by small uncharged residues; in bacteria predominantly Alanine. SPs of Gram-positive bacteria tend to be longer than those of Gram-negative bacteria.
Running signalP
SignalP takes amino acid sequences in fasta format. These are available from the annotation output as .faa files. Note that any letters not corresponding to the twenty standard amino acids, e.g. ‘U’, ‘B’, or ‘Z’, will be converted to ‘X’ and treated as unknown amino acids. Furthermore it is important to choose the correct organism group — Eukaryotes, Gram-negative bacteria, or Gram-positive bacteria with the -t option. The -m option will give a fasta output of the mature sequence (after cleavage of the signal peptide).
First, let’s make a new folder
$ cd ~/Secretome_prediction
$ mkdir prediction
Then, lets run signalP on the E.coli proteins
$ cd ~/Secretome_prediction/prediction
$ signalp -org gram- -mature -prefix Ecoli_secreted -fasta ~/Secretome_prediction/annotation/ERR022075/ERR022075.faa
where
-org string
Organism. Archaea: 'arch', Gram-positive: 'gram+', Gram-negative: 'gram-' or Eukarya: 'euk' (default "euk")
-prefix string
Output files prefix. (default "Input file prefix")
-fasta string
Input file in fasta format.
-mature
Make fasta file with mature sequences
Exercise
Generate a list of proteins with a signal peptide from the S.aureus genome.
Solution
$ cd ~/Secretome_prediction/prediction $ signalp -org gram+ -prefix Saureus_secreted -fasta ~/Secretome_prediction/annotation/S_aureus/S_aureus.faa
Alternatively, you can download the proteins to your own laptop and run them through the Signal-P server online.
Moving files between your instance and your laptop
Finally, we need to get results we produced to our own computers.There are also several ways to do this, but it’s always easier to start the transfer locally. This means if you’re using a transfer program, it needs to be installed on your local machine, not on your instance. If you’re typing into a terminal, the terminal should not be logged into your instance, it should be showing your local computer.
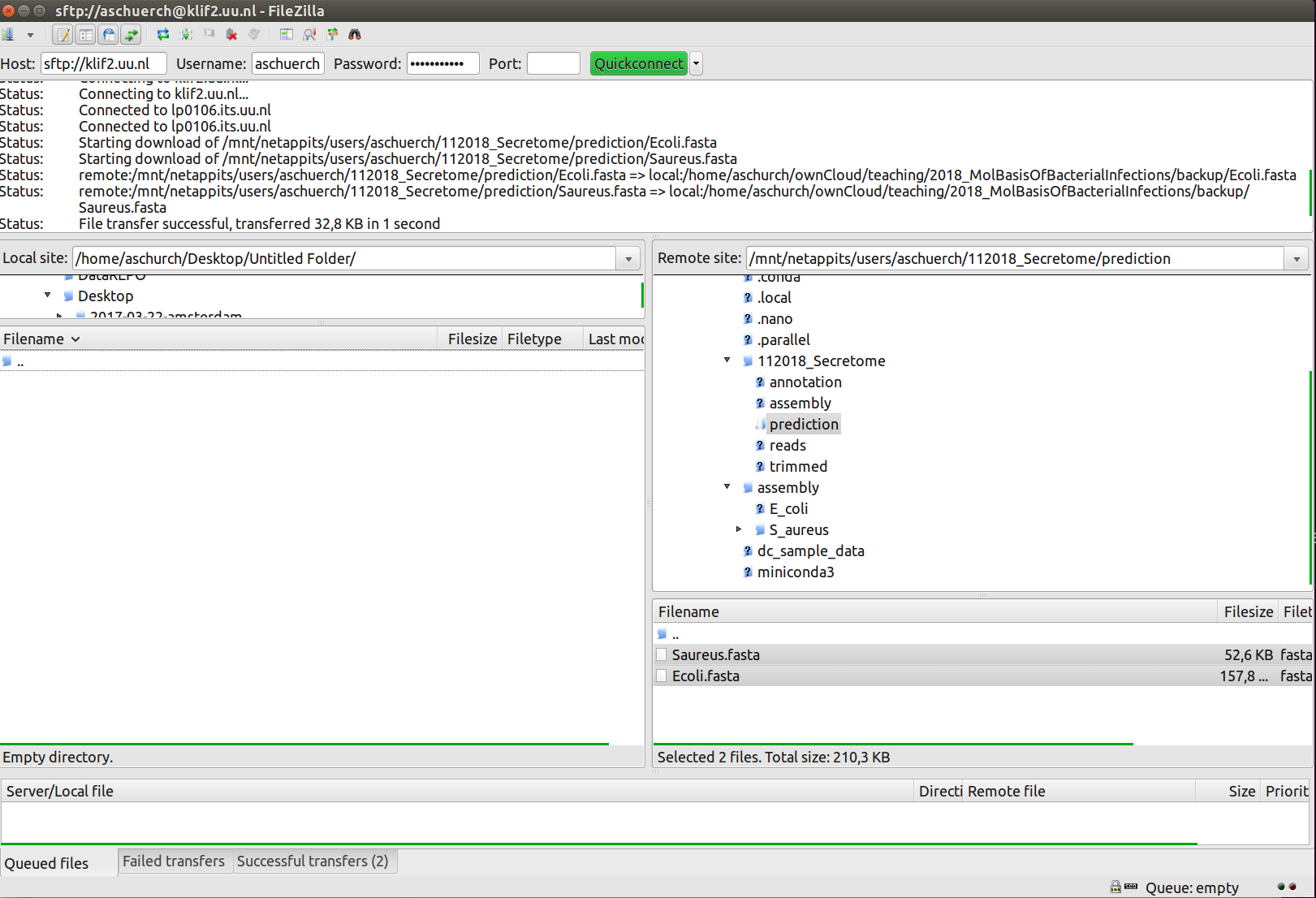
FileZilla is one of many free FTP clients that allow you to move files between local and remote locations.
It is convenient if you would like a graphical user interface to manage files and folders and if you have only a few files to move.
- Enter the location and user credentials of the remote server you are connecting to (upper panel)
- Enter your username and the password
- for port enter 22
- Files on your local computer will be in the highlighted (left panel)
- Your Desktop can be found in a folder called ‘Users’ > ‘your computer’s username’ > and Desktop
- Files on your remote computer will be in highlighted the right panel.
- Move files between computers by drawing and dropping them in the desired locations.
Tip: You can resize windows in FileZilla for easier viewing.
Key Points
Secreted proteins are predicted by identifying a signal peptide
The presence of a signal peptide does not necessarily mean that the protein is secreted
No matter which way you want to move data, it’s easier to start the transfer from your local machine