Popularity of SNP pipelines
Published:
Recently we got back a round of reviews on a small review paper. There was one sentence in the text were I listed some SNP pipelines that I thought were good examples. The topic of the review is not pipelines so the intention was not to list all, just one or two. However reviewer 2 did not agree with my choice of examples. He suggested to omit the name of the pipeline that was unpublished and name a few other published ones instead.
Well. In my mind it’s nice to have a reference but I do not have any problems of referencing a github page instead.
But of course the reviewer has a point. I just picked a few examples from my gut feeling without knowing what is really popular among microbiology laboratories. To get to the ground of it I set up a twitter poll, initially to get a feeling on what is used, and then a second survey on google docs where I tried to quantify the popularity of pipelines a bit more.
A total of 66 responses were recorded on the survey, with 50% of respondents in medical microbiology. Here is the very approximate map. Many people (24/66) indicated that they use several pipelines.
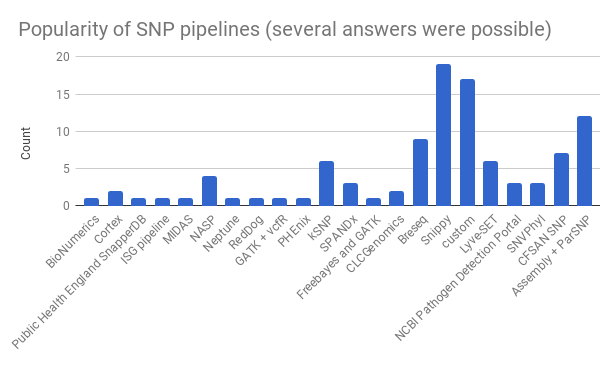
The most popular pipeline among respondents was Snippy with 19 mentions before custom pipelines which are used by 17. Also quite popular is the use of ParSNP on assembled genomes with 12 votes. This is slightly different from the earlier Twitter poll where 42% of 36 votes said to run a custom pipeline. The most striking thing is the diversity: there are 66 respondents and 21 different pipelines and that’s even without the custom pipelines.
Here’s a selection of reasons why the respondents use the pipeline they do:
- Same pipeline for eukaryotes and prokaryotes
- Quick!
- I wrote it
- At the moment when I needed it one of my co-workers used it
- Tested on dataset and found it was most accurate
- The others either limit parameter, are not updated often enough, or cannot be installed
- It works
- Easy to use
Of course, the population that took the survey is highly biased. The use of GUI pipelines for example seems to be extremely low, which would not be the case in the average European microbiology laboratory as far as I can tell.
Thanks everyone who participated in the survey.
You can browse through the all the responses here
