A list of plasmid prediction tools
Published:
In his recent publication Recovering Escherichia coli Plasmids in the Absence of Long-Read Sequencing Data Julian Paganini reviewed software tools to predict bacterial plasmids from sequencing data.
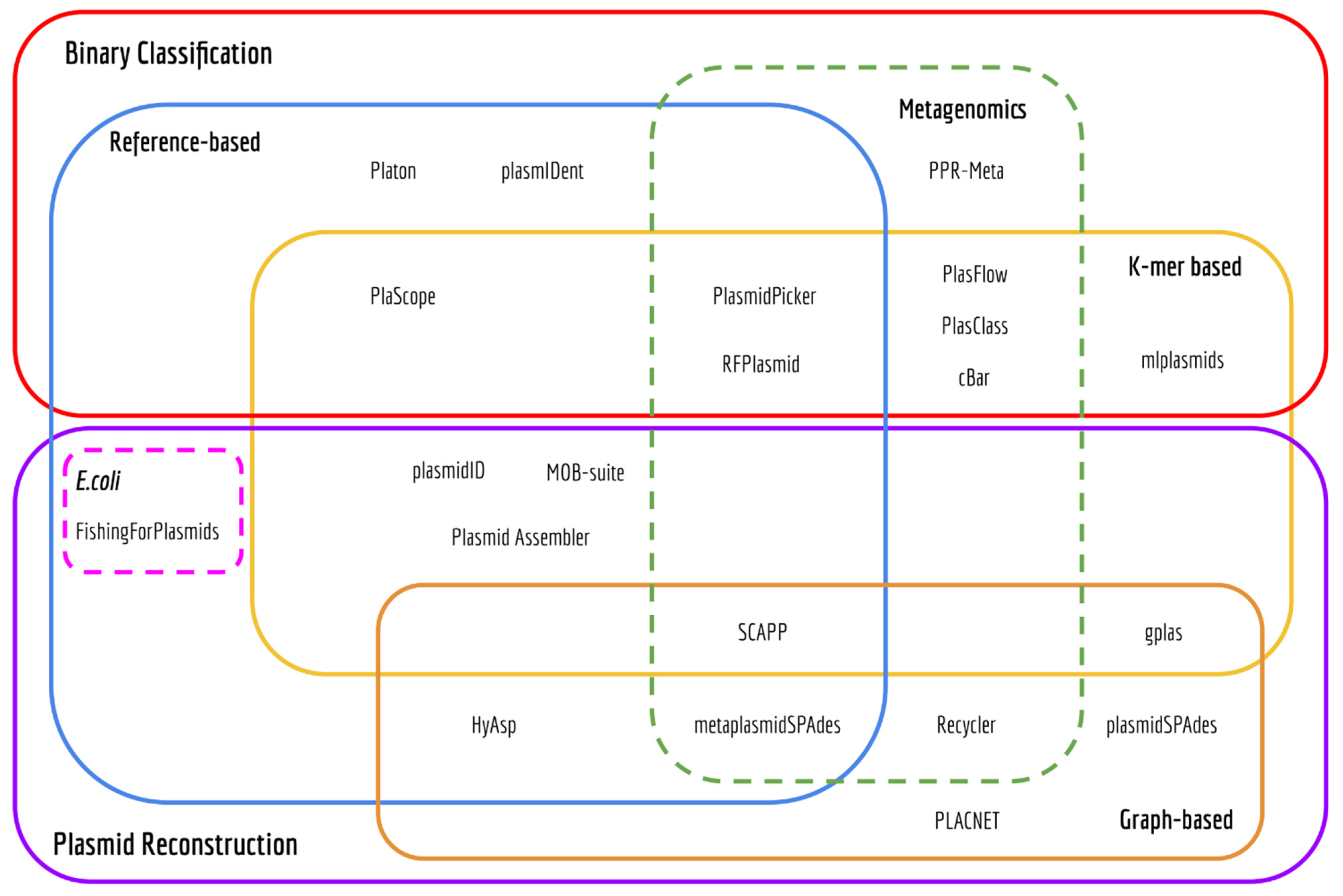
Here’s the list of all available tools (until 2020):
| Tools | Publication year | Input | Scope | Output | Reference DB | k-mer composition | Coverage | de Bruijn graph | plasmid-diagnostic sequences | strains meta-data | Length of contigs | “one-hot” matrix sequence representation | GC-content | Fully automatic? | User interface | Installation | Benchmarked in this study? | Retrieved from | Publication |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PlasmidSPAdes | 2016 | Reads | Multi-species | Plasmid bins | x | x | x | Command Line | Conda | yes | https://github.com/ablab/spades | https://doi.org/10.1093/bioinformatics/btw493 | |||||||
| metaplasmidSPAdes | 2019 | Reads | Meta-genomics | Plasmid bins | x | x | x | x | Command Line | Conda | no | https://github.com/ablab/spades | https://doi.org/10.1101/gr.241299.118 | ||||||
| HyAsp | 2019 | Reads or contigs and assembly graph | Multi-species | Plasmid bins | x | x | x | x | x | Command Line | Pip | yes | https://github.com/cchauve/HyAsP | https://doi.org/10.1093/bioinformatics/btz413 | |||||
| Recycler | 2017 | Assembly graph and BAM file | Multi-species/ Meta-genomics | Plasmid bins | x | x | x | Command Line | Conda | no | https://github.com/Shamir-Lab/Recycler | https://doi.org/10.1093/bioinformatics/btw651 | |||||||
| MOB-suite | 2018 | Contigs | Multi-species | Plasmid bins | x | x | x | x | Command Line | Conda | yes | https://github.com/phac-nml/mob-suite | https://doi.org/10.1099/mgen.0.000206 | ||||||
| gplas | 2020 | Assembly graphs | Multi-species | Plasmid bins | x | x | x | Command Line | Snakemake | yes | https://gitlab.com/sirarredondo/gplas/ | https://doi.org/10.1093/bioinformatics/btaa233 | |||||||
| FishingForPlasmids | 2020 | Contigs | E. coli | Plasmid bins | x | x | Command Line | Manual | yes | https://github.com/Visser-M/FishingForPlasmids | - | ||||||||
| PlasmidID | 2018 | Reads | Multi-species | Plasmid bins | x | x | x | x | Command Line | Conda | no | https://github.com/BU-ISCIII/plasmidID | - | ||||||
| Placnet | 2014 | Contigs, reads and BAM file | Multi-species | Plasmid bins | x | x | x | x | Command Line, GUI and Web-Application | SourceForge | no | https://github.com/LuisVielva/PLACNETw | https://doi.org/10.1093/bioinformatics/btx462 | ||||||
| SCAPP | 2020 | Assembly graph and reads | Meta-genomics | Plasmid bins | x | x | x | x | x | Command Line | Conda | yes | https://github.com/Shamir-Lab/SCAPP | https://doi.org/10.1101/2020.01.12.903252 | |||||
| Plasmid_Assembler | 2017 | Reads | Multi-species | Plasmid bins | x | x | x | x | Command Line | Conda | no | https://github.com/lowandrew/Plasmid_Assembler | - | ||||||
| PlaScope | 2018 | Contigs | E. coli | Binary classification | x | x | x | Command Line | Conda | no | https://github.com/labgem/PlaScope | https://doi.org/10.1099/mgen.0.000211 | |||||||
| PlasFlow | 2018 | Contigs | Meta-genomics | Binary classification | x | x | Command Line | Conda | no | https://github.com/smaegol/PlasFlow | https://doi.org/10.1093/nar/gkx1321 | ||||||||
| mlplasmids | 2018 | Contigs | Multi-species | Binary classification | x | x | Command Line and Web-Application | R-package | no | https://gitlab.com/sirarredondo/mlplasmids/ | https://doi.org/10.1099/mgen.0.000224 | ||||||||
| RFPlasmid | 2018 | Contigs | Multi-species/ Meta-genomics | Binary classification | x | x | x | x | Command Line and Web-Application | Pip | no | https://github.com/aldertzomer/RFPlasmid | https://doi.org/10.1101/2020.07.31.230631 | ||||||
| plasmIDent | 2018 | Contigs and long reads | Multi-species | Binary classification | x | x | x | x | Command Line | Nextflow / Docker | no | https://github.com/caspargross/plasmIDent | - | ||||||
| platon | 2020 | Contigs | Multi-species | Binary classification | x | x | x | x | Command Line | Conda | no | https://github.com/oschwengers/platon | https://doi.org/10.1099/mgen.0.000398 | ||||||
| PlasmidPicker | 2018 | Contigs | Meta-genomics | Binary classification | x | x | x | Command Line | Pip | no | https://github.com/haradama/PlasmidPicker | - | |||||||
| cBar | 2010 | Contigs | Meta-genomics | Binary classification | x | x | Command Line | Mannual | no | http://csbl.bmb.uga.edu/∼ffzhou/cBar | https://doi.org/10.1093/bioinformatics/btq299 | ||||||||
| PlasClass | 2020 | Contigs | Meta-genomics | Binary classification | x | x | Command Line | Auomatic set-up script | no | https://github.com/Shamir-Lab/PlasClass | https://doi.org/10.1371/journal.pcbi.1007781 | ||||||||
| PPR-Meta | 2019 | Contigs | Meta-genomics | Binary classification | x | x | Command Line | Manual | no | https://github.com/zhenchengfang/PPR-Meta | https://doi.org/10.1093/gigascience/giz066 | ||||||||
| PlasmidFinder | 2015 | Contigs | Multi-species | Reference plasmid detection | x | x | Command Line and Web-Application | Conda | no | https://bitbucket.org/genomicepidemiology/plasmidfinder/src/master/ | - | ||||||||
| PlasmidSeeker | 2018 | Reads and reference genome | Multi-species | Reference plasmid detection | x | x | x | x | Command Line | Manual | no | https://github.com/bioinfo-ut/PlasmidSeeker | https://doi.org/10.7717/peerj.4588 | ||||||
| PlasGUN | 2020 | Reads in fasta format | Meta-genomics | Plasmids gene prediction (other) | x | x | Command Line | Manual | no | https://github.com/zhenchengfang/PlasGUN/releases | https://doi.org/10.1093/bioinformatics/btaa103 | ||||||||
| plasmidtron | 2018 | Reads or contigs and metadata files | Multi-species | kmer based GWAS analysis of reads (other) | x | x | x | x | Command Line | Conda | no | https://github.com/sanger-pathogens/plasmidtron | https://doi.org/10.1099/mgen.0.000164 |
This table was published as Supplementary Table 1 and converted to Markdown with Table to Markdown.
Do you know any other tools that have not been included here? Let me know on Twitter.
